
Billions of dollars and countless hours of research have been invested toward developing preventative and therapeutic measures for cardiovascular disease. While enormous advances have been made, it remains the most common cause of death worldwide, claiming more than 16 million lives each year. A significant challenge is the fact that scientific inquiry and knowledge related to heart medicine remain distributed across many technical domains, without the benefit of a common language to unify them. For example, biological study at the molecular, cellular, tissue and organ levels have potential untapped synergies with development work on pharmaceuticals, medical devices, and clinical treatments.
One approach that offers promise to enhance this type of collaboration is to create computational models that draw from and contribute to multiple research disciplines. To that end, the Living Heart Project brings together specialists from academia, clinical practice, industry and regulatory bodies to create a common technology platform. This effort has produced the Living Heart Model (LHM), an anatomically accurate model for simulation and analysis of human heart function based on physiological principles. The underlying software engine for the model is the SIMULIA Abaqus® finite elements analysis product suite on the 3DEXPERIENCE platform.
Optimizing Performance to Enable Complex Modeling
The LHM generates the representation of an individual heart using data obtained from computed tomography (CT) and magnetic resonance imaging (MRI). The model includes all four chambers of the heart (left and right ventricles and atria), as well as all four valves (tricuspid, mitral, pulmonary, and aortic). This completeness results in far greater complexity than most prior models, which typically include only a subset of this anatomy.
Taking on the greater range of factors represented by all eight of these structures, however, allows for more complete modeling of phenomena such as the interplay between electrical stimulation and mechanical contraction of muscle tissue throughout the heart. Because irregularities in those relationships may be precursors to a heart attack or other dysfunction, modeling and analysis can provide vital contributions to better patient outcomes.
Adding further to the complexity of the LHM, it includes a number of sub-models, including a solid model, a finite element model, a muscle-fiber model and a fluid model. The solid model defines the physical anatomy of the heart, including chambers, vessels and other structures, while the muscle fiber model corresponds to the individual muscle sheets and fibers. The finite element model simulates dynamic electrical and mechanical behavior, and the fluid model represents the flow, pressure, and other characteristics of blood in the heart.
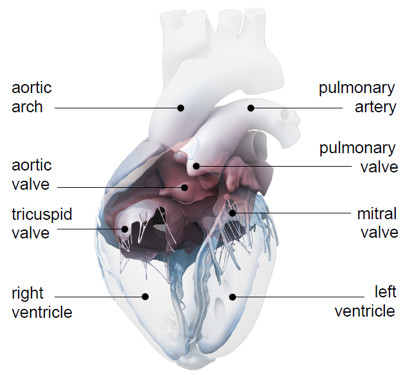
Internal anatomy of the heart model courtesy Zygote Media Group
The complexity of the LHM allows for very high-resolution representations of individual patients’ anatomical and physiological states. That lets researchers manipulate various model inputs and parameters to simulate the onset and progression of heart disease, helping them develop treatments that are as effective as possible for the needs of specific people.
As mentioned earlier, prior models were intentionally less complex than the LHM, as a matter of reducing computational complexity. Performance optimization of SIMULIA Abaqus for the Intel® Xeon® processor, carried out as part of an ongoing co-engineering relationship between Dassault Systèmes and Intel, helps address that challenge. In particular, Abaqus utilizes Intel® Advanced Vector Extensions 2.0 (Intel® AVX2) to accelerate computation with enhanced vectorization based on 256-bit registers, doubling the number of floating-point operations per cycle compared with the previous generation of the technology.
Steve Levine, LHP Director, expressed the value of this collaboration in real-world terms: “Computational Engineering and Biology are two of the most compute-intensive domains bottlenecking businesses today. The more than 35 percent performance improvements we’ve seen with the Intel Xeon processor E5-2697 v4 can directly translate into better products and reduced time to market for our customers. Further, with the addition of Intel AVX2, we’ve seen an additional performance boost. In the future, performance improvements will be measured in number of lives saved.”
In addition to that optimization at the per-core level—which ensures that individual threads execute efficiently—the Abaqus multiphysics simulation and finite-element analysis software that underlie the LHM are also tuned to take advantage of large-scale computing resources. Intel and Dassault Systèmes have collaborated to ensure that the software divides workloads effectively among execution cores at the node level, and that code operates efficiently across multiple nodes at the cluster level.
The Endeavour cluster, located at the Customer Response Team data center operated by Intel in Rio Rancho, New Mexico, provides optimized infrastructure based on Intel Xeon processors and Intel® Xeon Phi™ coprocessors for the development and operation of the LHM. A Top500 Supercomputing site, Endeavour is used primarily for benchmarking in the high-performance computing market segment. The cluster includes more than 12,000 cores in total, of which hundreds were reserved and made available for the many hours of time needed to prepare, tune, and execute LHM workloads. These resources were instrumental in enabling the LHM to accurately simulate and analyze heart function, including three-dimensional structure, electromechanics, and fluid dynamics.
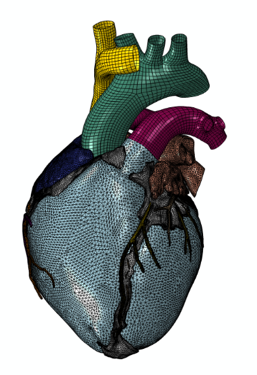
Finite Element representation
A Novel, Dual-Resolution Approach to Acceleration
Even with the support of performance optimizations and extensive compute resources to accelerate computations for the LHM, simulations remain computationally intensive, and they often take several hours or even days. This time requirement can often be prohibitive, limiting the value of the LHM in various usages. In clinical environments, delays can hamper the ability to make decisions that affect the progress and even outcome of treatment. In the design of medical devices, longer time to completion can interfere with product development, which can have financial consequences. In virtual clinical trials, long-running simulation cycles can limit the number of iterations that can be made, potentially impacting the success of a study.
Researchers from Dassault Systèmes, Intel, and the Massachusetts Institute of Technology (MIT) envisioned a novel way of reducing the time requirements to run high-resolution simulation models such as the LHM. That is, the team hypothesized that a fast-running low-resolution heart model could be used to provide precise inputs to the longer-running high-resolution model. By predicting how the high-fidelity model would behave over time, the low-fidelity model could be used as a calibration mechanism, reducing overall compute time.
To test this approach, the team created a low-fidelity lumped-parameter model (LPM) made up of three passive chambers, blood vessels, and a pulmonary-circulation representation, matched to the LHM’s circulatory model. After an initial set of training runs on LHM to simulate a baseline and various disease states, a set of runs with the LPM (which required less than one second of execution time each) generated predictions of the LHM end pressure/volume state. The outputs of the fast-running LPM were then inputted as initial parameters of the long-running LHM, for both the baseline state and the disease states.
Coupling the high-fidelity model with the low-fidelity one reduced computation time to reach steady state from 20 hours to four hours compared to the high-fidelity model alone, when running on 24 cores. This dramatic acceleration has led the team to ongoing work, calibrating the combined model to represent a broad range of physiological and pathophysiological states. The hope is to enable insight across research, clinical, and regulatory domains.
Ketan Paranjape, Intel General Manager for Life Sciences and Analytics, expressed his optimism: “The Living Heart Project extends the value of high-performance computing in the life sciences, alongside research and therapies in areas such as genomics, pharmacology, and computational biology. Intel is committed to applying our computing innovation to advance human knowledge across scientific disciplines, and it’s immensely satisfying when we see these efforts contribute to the greater human good.”
Moving from the Lab to Patient Practice
While the vast array of interventions and other choices available to cardiac doctors and patients is an enormous benefit, the range of choices adds enormous complexity to treatment. A key goal of the LHP is to facilitate better choice making in this area, as a means of improving patient care. To that end, studies are already underway that simulate heart disease and the potential effects of drugs and medical devices under various conditions. In the future, that approach could potentially be expanded to act in conjunction with models of other human systems, beyond the cardiovascular system.
Other non-surgical techniques often provide only clues as to what the future might hold for a given patient after receiving a given course of treatment. As an alternative, the LHP represents a step toward more concrete prediction and decision-making. In addition to improving outcomes, treatment based on sophisticated computational modeling such as the LHP may represent an approach that is both less invasive and less expensive than alternatives.
Researchers look ahead to a host of potential ways that LHM could contribute to patient care. Beyond the ability to provide robust simulations of individualized disease states, the technology can be paired with three-dimensional rendering to create compelling visualizations. For example, showing a patient an immersive, interactive representation of her beating heart in clinic could make it easier to explain a cardiovascular condition and aid the doctor and patient as they consider treatment options.

Computational models of the human heart enable robust 3D visualizations for use in clinical, educational, and research settings.
Conclusion
Ongoing research on the LHM highlights cardiac research and clinical care as examples of joint work by diverse, complementary disciplines creating a whole that is greater than the sum of its parts. The project demonstrates how high-performance computing using standards-based hardware can foster new dimensions of collaboration by academia, industry, and others for mutual benefit and advancement.
Innovations such as accelerating simulations using the dual-resolution approach developed by Dassault Systèmes, Intel, and MIT advance potential for solutions beyond what is apparently possible with current levels of compute power. That spirit of expanding reality is behind all invention, and in the case of LHP, it also has the potential to improve the quality of life and prognosis for millions of people.
Matt Gillespie is a technology writer based in Chicago. He can be found at www.linkedin.com/in/mgillespie1.




