A team from The University of Texas at Austin, the University of Pennsylvania, the University of Houston and the University of Stuttgart competed in the 2017 BRaTS Challenge using advanced computing resources at the Texas Advanced Computing Center. Credit: TACC
Researchers are turning to advanced techniques to better identify brain tumors.
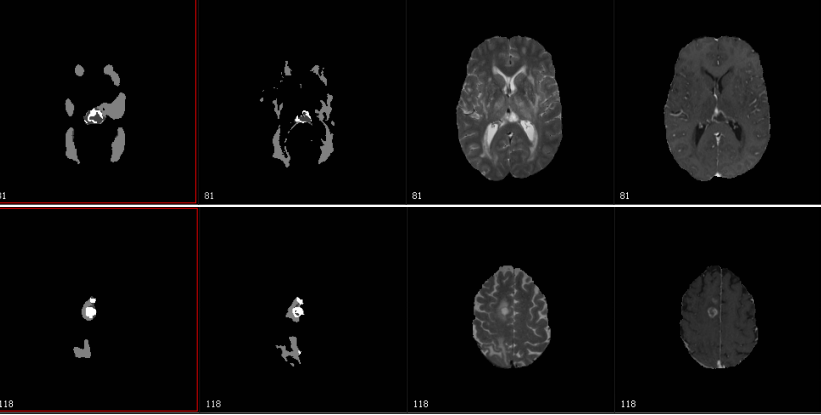
During the 20th International Conference on Medical Image Computing and Computer Assisted Intervention Sept 10-14, a research group from the University of Texas at Austin’s Texas Advanced Computing Center (TACC) presented results of a new, fully automatic method combining biophysical models of tumor growth with machine learning algorithms for the analysis of Magnetic Resonance (MR) imaging data of glioma patients.
The research team, led by George Biros, professor of mechanical engineering and leader of the ICES Parallel Algorithms for Data Analysis and Simulation Group at Texas, tested the method in the Multimodal Brain Tumor Segmentation Challenge 2017—an annual competition where research groups from around the world present methods and results for computer-aided identification and classification of brain tumors and other types of cancerous regions using pre-operative MR scans.
Biros’s group landed in the top 25 percent of the challenge.
“The competition is related to the characterization of abnormal tissue on patients who suffer from glioma tumors, the most prevalent form of primary brain tumor,” Biros said in a statement. “Our goal is to take an image and delineate it automatically and identify different types of abnormal tissue—edema, enhancing tumor [areas with very aggressive tumors] and necrotic tissue.
“It’s similar to taking a picture of one’s family and doing facial recognition to identify each member but here you do tissue recognition, and all this has to be done automatically,” he added.
Primary brain tumors encompass a wide range of tumors depending on the cell type, the aggressiveness and the stage of the tumor.
Currently, a trained radiologist must accurately characterize the scope of the tumor, but advanced computer technologies could assist the professional in accelerating the assessment.
During the challenge, the researchers were provided with 300 sets of brain images in advance where all teams calibrated their methods.
They then were given data from 140 patients and were asked to identify the location of tumors and segment them into different tissue types of the over the course of two days.
The method they used included a supervised machine learning step where the computer creates a probability map for the target classes and a second step where they combine the probabilities with a biophysical model that represents how tumors grow in mathematical terms.
TACC computing resources enabled the team to use large-scale nearest neighbor classifiers—where for every voxel or 3D pixel, in a MR brain image, the system tries to find all the similar voxels in the brains it has already seen to determine if the area represents a tumor or a non-tumor.
The team was able to analyze the pipeline on 140 brains in less than four hours and correctly characterize the testing data with nearly 90 percent accuracy.
The researchers now plan on using the tools and basic ideas to scale up, while also improving the technique.




