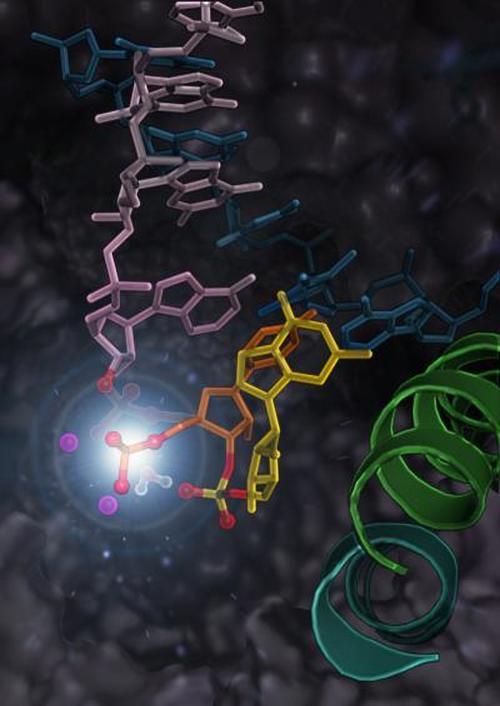
RNA polymerase II undergoes the intrinsic cleavage of the mis-incorporated nucleotide (the yellow part in the picture) during proofreading of the RNA transcription. (Credit: The Hong Kong University of Science and Technology)
The message of life is encoded in our genomic DNA through transcription of messenger RNAs and translation of proteins to perform cellular functions. To ensure accurate transcription—a process that transcribes genomic DNA into messenger RNA by adding nucleotides one by one like letters in the alphabet, an enzyme called RNA polymerase II would synthesize and proofread messenger RNA to remove any mis-incorporated nucleotides that do not match with the DNA template.
While RNA polymerase II was known to be critical in ensuring the accuracy of transcription, it had been a long-standing puzzle as to how this enzyme accomplishes this difficult task. Scientists have always been intrigued to find out the underlying mechanisms, as that could offer insights on how errors could be made during this otherwise highly-accurate transcription process, which may lead to various human diseases.
A research team led by Prof. Huang Xuhui, Padma Harilela Associate Professor of Science in the Department of Chemistry and Department of Chemical and Biological Engineering at HKUST, recently discovered the mechanism for RNA polymerase II to correct errors in RNA synthesis. When a nucleotide is added by mistake, RNA polymerase II can rewind by moving backwards (called backtracking) and cleave this mis-incorporated nucleotide. The research team found that while specific amino acid residues of RNA polymerase II are critical for backtracking, cleavage of the mis-incorporated nucleotide only requires the RNA itself (i.e. phosphate oxygen of mis-incorporated nucleotide).
“RNA polymerase II is like a molecular machine in the cell. Nature cleverly designs this machine to catalyze two distinct chemical reactions in a single active site without getting mixed-up. While normal RNA synthesis requires specific amino acid residues of RNA polymerase II, we found that the removal of the mismatched nucleotide does not rely on any amino acid residues. This molecular machine seamlessly coordinates these two functions in one active site,” said Huang. “Our discovery offers valuable insights into how transcription may go wrong in ageing and diseased cells, and to what extent transcriptional errors may lead to various human diseases.”
“Our work is only possible with the large-scale high-performance computing resources mostly provided by the Shaheen Supercomputer in collaboration with King Abdullah University of Science and Technology (KAUST),” Huang added. “Our quantum mechanics and molecular dynamics calculations consumed 20 million CPU core hours in total.”
The findings were recently published in scientific journal Nature Catalysis.
Huang’s research interest lies in understanding complex biological and chemical processes using computational approaches. The computational work was conducted by Dr. Carmen TSE, a long-time HKUST affiliate who completed her undergraduate, Ph.D., and post-doctoral training all at HKUST’s Department of Chemistry. Other corresponding authors include HKUST’s Research Assistant Professor Dr. Peter Cheung from Department of Chemistry, Prof. Yingkai Zhang from New York University, and Prof. Wang Dong from University of California, San Diego whose research group performed experiments to validate the predicted mechanisms from calculations.



