While the dawn of ChatGPT in 2022 helped kickstart the current AI wave, an AI moment that arguably mattered more for science was AlphaFold. December 2018, first place at CASP13, the Critical Assessment of Structure Prediction competition. By 2020, protein structure prediction was close enough to “solved” that the real question shifted from whether a fold was possible to what you could build on top of it. By 2024, it was Nobel Prize territory.
AlphaFold spawned an ecosystem: open-source competitors, specialized forks, commercial platforms built on its architecture. But it also helped legitimize a broader shift. Astronomers started using neural networks to classify galaxies faster than graduate students ever could. Climate modelers trained AI on decades of satellite data to sharpen precipitation forecasts. Epidemiologists built models that could track disease variants in near real-time. Drug developers stopped asking whether AI belonged in the pipeline and started asking which parts of the pipeline it could own.
Now in 2025, across disciplines, software is increasingly shaping which experiments get run. In structure prediction alone, the year delivered a wave of tools that built directly on AlphaFold’s foundation.
1. Protein structure prediction moves from folding to binding
The 2025 generation of structure prediction tools solved a problem AlphaFold left open: predicting how tightly a drug candidate will bind to its target. Boltz-2, released in June by MIT CSAIL and Recursion, predicts structure and binding affinity jointly, running 1,000 times faster than physics-based free energy perturbation methods. Genesis Molecular AI’s Pearl, backed by NVIDIA, claimed 40% improvement over AlphaFold 3 on drug discovery benchmarks and provided the first evidence of scaling laws in molecular AI. Meanwhile, the OpenFold Consortium released OpenFold3 in October.
The David Baker Lab at University of Washington pushed the design frontier. RFdiffusion2 (April) designs enzymes from functional group geometries. RFdiffusion3 (December) runs 10 times faster with atom-level precision. MIT’s BoltzGen, released in November, generates protein binders from scratch, achieving nanomolar binding for 66% of novel targets tested.
Boltz-2 image as shown on an NVIDIA website
Major releases in structure prediction
| Tool | Date | Developer | What it does | Why it matters | Sources |
|---|---|---|---|---|---|
| Boltz-2 | June 2025 | MIT CSAIL / Recursion | Predicts structure + binding affinity jointly | 1000x faster than physics-based FEP; open-source under MIT license; already in Recursion’s pipeline | Recursion PR ∙ GEN ∙ C&EN |
| BoltzGen | November 2025 | MIT | Generative AI for designing protein binders from scratch | Targets hard-to-treat disease targets; de novo molecule creation; nanomolar binders for 66% of novel targets | MIT News ∙ bioRxiv ∙ GEN |
| OpenFold3 | October 2025 | OpenFold Consortium | Open-source AlphaFold 3 competitor | Approaching AF3 parity; community-driven development | Nature |
| Pearl | October 2025 | Genesis Molecular AI | Structure prediction optimized for drug discovery | Claims up to 40% improvement over AlphaFold 3 on key benchmarks; first evidence of scaling laws in molecular AI; NVIDIA collaboration | Business Wire ∙ Genesis ∙ MIT Tech Review |
| RoseTTAFold Diffusion 2/3 | 2025 | David Baker Lab / UW | Protein design via diffusion models | RFdiffusion2 (April 2025): designs enzymes from functional group geometries; RFdiffusion3 (Dec 2025): 10x faster, atom-level precision | Nature Methods ∙ Chemistry World ∙ IPD |
| Schrödinger FEP+ Protocol Builder | 2023 (active 2025) | Schrödinger | Active learning for free energy perturbation protocol optimization | Automates what was a manual, expertise-driven process; generates optimized models 4x faster than manual | Schrödinger ∙ J Chem Inf Model |
Structure prediction has clean edges: sequence in, coordinates out, benchmark against experiment. The 2025 bet from frontier labs was whether the same logic could scale to messier problems—literature synthesis, experimental design, the part of research that lives in docs and Slack threads before it reaches a protocol.
2. Frontier labs build AI research assistants with real integrations
One of the most influential players in science AI is Google, whose researchers helped create both AlphaFold and the transformer architecture underlying most modern language models. In February 2025, Google unveiled AI Co-Scientist, a multi-agent system built on Gemini 2.0 (now a full generation behind the company’s latest LLM model) that generates hypotheses, designs experiments, and drafts research proposals. In one validation test, the system independently arrived at a bacterial gene transfer mechanism that Imperial College researchers had spent a decade confirming, in 48 hours.
Google is not alone in targeting the research workflow. OpenAI launched Deep Research the same month, a tool that synthesizes hundreds of scientific papers into cited reports in under an hour, essentially automating the literature review that consumes weeks of a PhD student’s life. Anthropic followed in October with Claude for Life Sciences, the company’s first formal scientific product, featuring direct integrations with Benchling, PubMed, 10x Genomics, and BioRender. At Novo Nordisk, the platform cut clinical study documentation from over 10 weeks to 10 minutes.
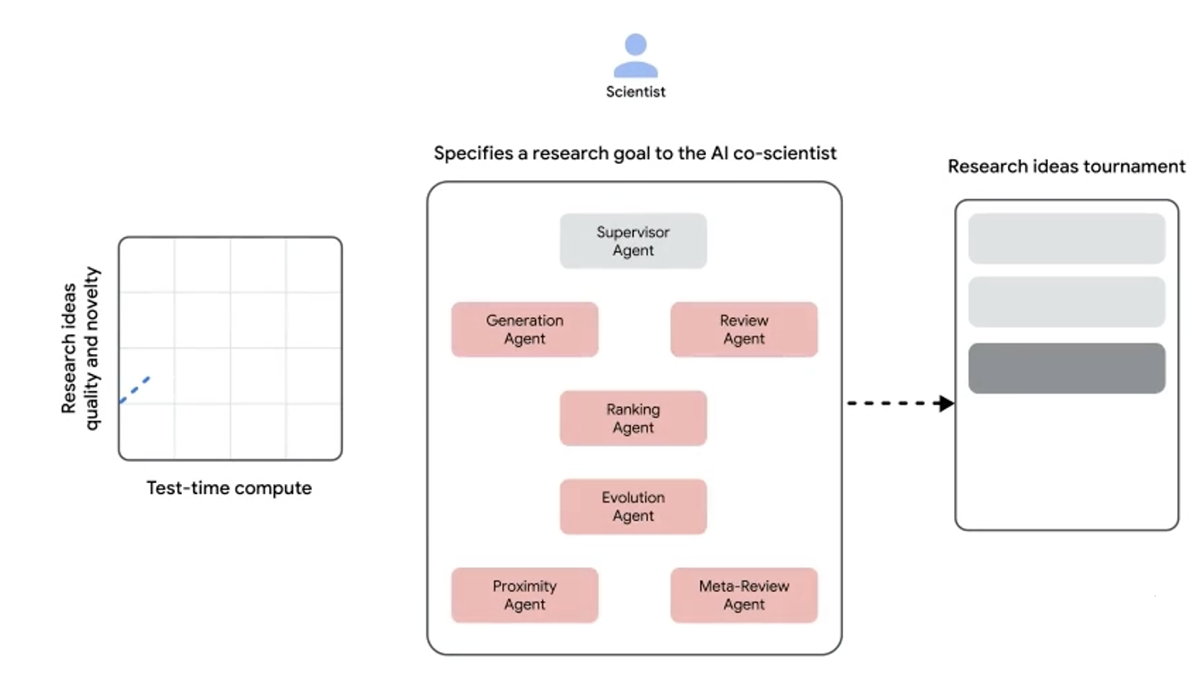
A schematic of AI co-scientist as shown on a Google blog
Major releases
| Tool | Date | Developer | What it does | Why it matters | Sources |
|---|---|---|---|---|---|
| Google AI Co-Scientist | February 2025 | Google DeepMind | Multi-agent system on Gemini 2.0; generates hypotheses, designs experiments, drafts proposals | Virtual research collaborator, not just an assistant; independently replicated decade-long research in 48 hours | Google Research ∙ arXiv ∙ HPCwire |
| OpenAI Deep Research | February 2025 | OpenAI | Synthesizes hundreds of papers in <1 hour with audit trails | “AI PhD student” concept becomes tangible; powered by o3 model optimized for browsing/analysis | OpenAI ∙ Nature |
| OpenAI FrontierScience Benchmark | December 2025 | OpenAI | Tests AI on expert-level reasoning in physics, biology, chemistry | 700+ questions; Olympiad and Research tracks; GPT-5.2 scores 77% Olympiad, 25% Research | OpenAI ∙ TIME |
| Claude for Life Sciences | October 2025 | Anthropic | Connectors to Benchling, PubMed, 10x Genomics, BioRender; skills for scRNA-seq QC | First formal life sciences product from a frontier lab | Anthropic ∙ CNBC |
| Anthropic AI for Science Program | May 2025 | Anthropic | Up to $20K API credits for qualified researchers | Democratizes access for academic labs | TechCrunch ∙ Anthropic Support |
Anthropic’s Benchling connector only works if your Benchling is populated. Google’s AI Co-Scientist can draft a proposal, but someone still has to know where the samples are. The bottleneck isn’t the model—it’s the metadata.
3. Lab informatics platforms add AI to core workflows
Laboratory information management systems (LIMS), electronic lab notebooks (ELNs), and inventory systems received significant upgrades in 2025. The difference from previous years: AI features moved from marketing bullet points to functionality that changes daily workflows. LabVantage 8.9 introduced voice command support through its Lottie/Open Talk system, unveiled at PittCon 2025. Sapio Sciences launched what it calls an “AI Lab Notebook” with molecular docking and ad hoc analytics built into the interface, demonstrated at SLAS 2025. Scispot positioned its LabOS as an API-first “Lab Operating System” connecting to 7,000+ applications.
Major releases
| Tool | Date | Developer | What it does | Why it matters | Sources |
|---|---|---|---|---|---|
| LabVantage 8.9 | March 2025 | LabVantage Solutions | Voice command support (Lottie/Open Talk), AI-driven efficiency, database partitioning for speed | “SaaS 2.0” positioning; agentic AI integrations; unveiled at PittCon 2025 | LabVantage PR ∙ Business Wire |
| LabWare Assure | April 2025 | LabWare | New SaaS portfolio addition (joins GROW and QAQC) | Food safety and quality LIMS; expands cloud options for regulated environments; announced at PittCon 2025 | PR Newswire ∙ LabWare |
| InstantGMP LIMS | October 2025 | InstantGMP | Integrated QC module built into manufacturing platform | Purpose-built for GMP/GLP labs; unifies QC with production/inventory; Part 11 compliant | PR Newswire ∙ InstantGMP |
| Sapio ELaiN | September 2024 (GA); September 2025 (“3rd-gen ELN”) | Sapio Sciences | AI-native lab assistant; natural language data queries; “agentic” AI | First AI Lab Notebook (AILN); demonstrated at SLAS 2025; molecular docking, ad hoc analytics built-in | Sapio Sciences ∙ SLAS Blog |
| Scispot LabOS | Active 2025 | Scispot | Unified ELN + LIMS + SDMS with AI, Jupyter integration | API-first “Lab Operating System” for biotech; no-code configuration; connects 7,000+ apps | Scispot ∙ LabOS blog |
BayBE can pick your next experiment. It cannot pick your next experiment if it doesn’t know what you just ran, what failed, and what’s left in the freezer. That’s why LIMS upgrades and autonomous labs show up in the same year: one enables the other.
4. Self-driving labs move from demos to deployment
In 2025, autonomous labs moved from academic proof-of-concept to scaled deployment. A few products followed the footsteps of systems like Argonne National Laboratory’ Polybot, which launched in 2023. For instance, Lawrence Berkeley National Lab’s Distiller, released in April, streams electron microscope data to the Perlmutter supercomputer for real-time analysis. NC State’s Abolhasani lab published techniques in July that enable 10 times more data collection per experimental run through flow-driven data intensification.
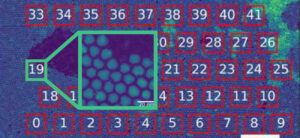
Automated atomic-resolution imaging of core-shell nanoparticles, with a low-magnification view highlighting regions of interest and an atomic-scale inset of one selected area. Credit: Berkeley Lab.
The enabling software is maturing alongside the hardware. BayBE, an open-source Bayesian optimization framework from Merck KGaA and the University of Toronto Acceleration Consortium, has become core infrastructure for closed-loop experimentation. Sakana AI’s “AI Scientist,” released in August 2024 with collaborators from Oxford and UBC, demonstrated a complete autonomous research loop from idea generation through code, experiments, and paper drafting.
McKinsey estimated in 2023 that comprehensive automation could cut pharma R&D costs by approximately 25%. The pattern across these deployments: closed-loop systems connecting AI planners to robotic execution to automated analysis, with emphasis on resource savings and iteration speed.
Major developments
| Tool/Initiative | Date | Developer | What it does | Why it matters | Sources |
|---|---|---|---|---|---|
| BayBE | December 2023 (active 2025) | Merck KGaA / U of Toronto Acceleration Consortium | Open-source Bayesian optimization for closed-loop experimentation | “Brain” for automated equipment; Apache 2.0 license; powers dozens of Merck use cases | Merck PR ∙ U of Toronto ∙ GitHub |
| The AI Scientist | August 2024 | Sakana AI / U of Oxford / UBC | Autonomous research loop: idea → code → experiment → paper | Proof of concept for end-to-end AI research; ~$15/paper; open source | Sakana AI ∙ arXiv ∙ GitHub |
| NC State SDL techniques | July 2025 | NC State (Abolhasani lab) | Flow-driven data intensification for 10x data collection at record speeds | Slashes costs, waste; Nature Chem Eng paper; “Rainbow” multi-robot SDL (Aug 2025) | NC State News ∙ ScienceDaily |
| Argonne Polybot / Autonomous Discovery | February 2025 | Argonne National Lab | AI-driven automated materials laboratory; autonomous polymer thin-film processing | Federal lab investment in SDLs; Nature Communications paper; part of DOE Genesis Mission | Argonne ∙ UChicago News |
| Berkeley Lab Distiller | April 2025 | Lawrence Berkeley National Lab | Web-based platform streaming electron microscope data to Perlmutter for real-time analysis | Enables experiment refinement while still running; DOE Superfacility concept | Berkeley Lab ∙ NERSC |
NC State’s SDL collects 10x more data per run than previous setups. Berkeley’s 4D Camera generates 480 gigabits per second. Autonomy solves the throughput problem and creates the analysis problem. In genomics, that lands on bioinformatics.
5. Genomics data infrastructure scales for multiomics
The pipes that move genomic data from instruments to insights received critical upgrades in 2025. Illumina’s DRAGEN v4.4, released in May, delivered a 30% improvement in structural variant calling alongside oncology applications, multiomics support, and AWS F2 instance availability. The platform’s built-in AI/ML for variant calling has made it the industry standard for NGS secondary analysis.
On the corporate side, QIAGEN acquired Parse Biosciences in November for $225 million upfront plus $55 million in milestones, expanding its single-cell sequencing and bioinformatics capabilities. Parse’s Evercode platform serves more than 3,000 labs. The acquisition reflects a broader consolidation trend as vendors build end-to-end workflows from sample prep through analysis.
Major releases
| Tool | Date | Developer | What it does | Why it matters | Sources |
|---|---|---|---|---|---|
| Illumina DRAGEN v4.4 | May 2025 | Illumina | 30% improvement in SV calling; oncology apps; multiomics support; AWS F2 instances | Industry-leading NGS secondary analysis; built-in AI/ML for variant calling | PR Newswire ∙ Illumina |
| QIAGEN acquires Parse Biosciences | November 2025 | QIAGEN | Expands single-cell sequencing + bioinformatics | $225M upfront + $55M milestones; Parse’s Evercode platform serves 3,000+ labs | QIAGEN PR ∙ Business Wire |
6. Open-source scientific tools gain institutional backing
Press releases announce features. GitHub commits show who’s maintaining them. For a field where vendor claims outpace validation, public repositories are one of the few places you can check the work.
The Python scientific ecosystem continued maturing in 2025, with several packages crossing from “researcher tools” to “infrastructure.” The Boltz repository (jwohlwend/boltz) grew to more than 1,300 Slack community members and 200+ biotech adopters. OpenFold3 is approaching AlphaFold 3 parity with community-driven development and backing. Merck’s BayBE has become core to the company’s self-driving lab strategy while remaining fully open-source (also on GitHub).
Longer-standing tools maintained their central roles: scanpy remains the a common tool for single-cell RNA-seq analysis, and RDKit continues as the backbone of computational chemistry. AlphaFold itself has accumulated more than 43,000 citations, with weights released in November 2024.
Key repositories
| Repo | What it is | 2025 status | Link |
|---|---|---|---|
jwohlwend/boltz |
Boltz-1/2 protein structure + affinity | >1,300 Slack community; 200+ biotech adopters | GitHub |
aqlaboratory/openfold |
Open-source AlphaFold | OpenFold3 approaching AF3 parity | GitHub |
Merck/BayBE |
Bayesian experiment optimization | Core to Merck’s self-driving lab strategy | GitHub |
scverse/scanpy |
Single-cell analysis | Workhorse of scRNA-seq field | GitHub |
rdkit/rdkit |
Cheminformatics toolkit | Backbone of computational chemistry | GitHub |
google-deepmind/alphafold |
AlphaFold 2/3 | 43K+ citations; weights released Nov 2024 | GitHub |




Tell Us What You Think!
You must be logged in to post a comment.