Written by Peter Christey, PhD, co-founder and CEO of General Automation Lab Technologies Inc.
A nanoscale array-based format could facilitate high-throughput cultivation of individual microbial strains.
Thanks to exciting new insights into the nature and importance of microbiomes, the field of microbiology is being reinvigorated. Indeed, significant R&D resources are being committed to realizing the promise of microbiome-based diagnostics and therapeutics. Scientific papers consistently report advances from microbial studies, whether it’s improving our understanding of microbial communities in the environment or establishing associations between diseases and microbiomes in the human body.
But even as we celebrate the promise of microbiome-based advances, the sad reality is that this field remains limited by century-old microbiology tools. Too many microbial studies still depend on conventional culture techniques, which are prohibitively low-throughput for the kinds of large-scale studies needed today. In addition to the time and labor they require, culture methods also introduce significant bias because the most abundant or fastest-growing microbial strains outcompete their rarer or slower-growing counterparts. Downstream analyses may not even detect the presence of these less visible—but still biologically important—microbes in the community being studied.
To get around the limitations of current culture methods, many scientists use next-generation sequencing (NGS) platforms to study microbial communities. This has been quite successful for generating data about the genomes or transcriptomes of these populations. However, a true understanding of what is going on in a microbiome necessitates far more than cataloguing the strains present in a sample; we must also embrace the full complexity of these communities by analyzing the functions and interactions of their constituents. For example, while DNA can offer a look at the population differences between a microbiome associated with health and one associated with disease, it cannot reveal the underlying biological mechanisms that might establish causation between the microbiome and host health. The in-depth studies needed to elucidate function and interaction must be done on living communities and isolates, not the processed libraries used for NGS pipelines.
There are similar limitations with other ’omics approaches, such as metabolomics or proteomics. What the scientific community needs is an entirely new approach to culturing microbes — one that would enable the study of living organisms, but that would also support the kinds of high-throughput analyses demanded by modern science. The ideal solution would be automated to reduce hands-on time, high-capacity to support the growth of thousands of microbial strains at a time, and economical to make large-scale studies feasible.
Microbiome Successes
While we are still in the early stages of exploring and making sense of microbiomes, there have already been major advances in how we appreciate the ways that these communities influence our lives and environment.
For example, microbiome research in agriculture is transforming our view of crop growth, yield, drought resistance, and pest resistance. Scientists have determined that the plant domestication process itself may have diminished the microbial communities living in and around crops, reducing the microbes’ ability to promote growth and health1. While follow-on functional studies must be done to more fully characterize the relationship between crop plants and microbial populations, these results offer hope that adjusting microbiomes could be an effective way of improving crop yield without increasing the land area allocated to farming.
There is even some preliminary data to suggest that fine-tuning microbial populations in soil may help alleviate the effects of climate change2. Microorganisms have important functions in carbon and nutrient cycling, and deploying them to remove carbon from the atmosphere and store it in soil is an approach that some researchers believe could have global benefits.
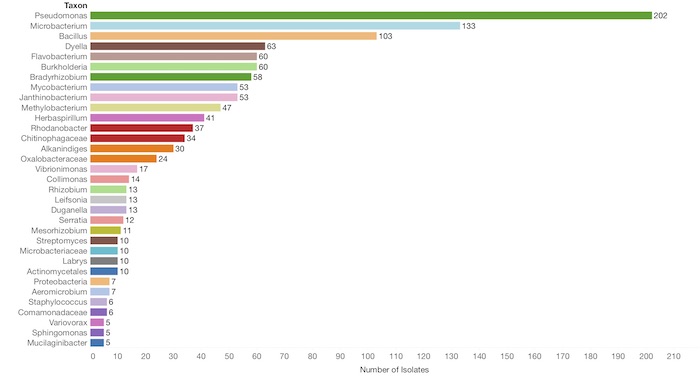
Data from an experiment that generated taxonomic identification of 1,251 isolates from an urban soil sample.
Perhaps the most intriguing results from microbiome studies to date have come from associations between the microbes we carry in our bodies and the states of our health. Several studies to date have shown that microbial populations may affect the progression of a tumor, and also that microbes help shape a person’s response to cancer immunotherapy3,4. Scientists involved in these projects have called for further analysis to prove the functional connection between microbes and cancer progression or treatment response. Of course, the microbiome has been implicated in many other health conditions as well: autism, inflammatory bowel disease, type 2 diabetes, and preterm birth are just a few.
Technical Challenges
Despite this progress, limitations with current microbiology tools are preventing scientists from accomplishing even more. The Petri dishes and agar plates used for cultivating microbes have been mainstays in laboratories for more than a century. They work well with fast-growing or populous microbes, but are not very successful with slow-growing strains or rare species in a population.
Most of the tools available to microbiologists today were developed in an era when the primary goal was either to detect the presence of microbes or to identify a single strain. They were not meant to address the goal of so many researchers today: to separate, identify, and comprehensively characterize hundreds or thousands of strains living together in a complex community.
Because culturing is so time-intensive and has inherent bias, many scientists elect to skip this step and analyze microbial populations with high-throughput NGS platforms. While this has been very helpful for cataloguing species, the approach is unable to provide critical information about microbial functions—individually and collectively—and their interactions. These are both important dimensions for establishing causal connections between microbiomes and their environment or host, but it is not possible to study either without starting with live organisms.
Researchers have also adopted proteomic approaches for characterizing microbial communities, but the mass spectrometry-based methods used for this cannot accurately associate each peptide identified to the specific microbial strain from which it came. It is also very difficult to analyze novel proteins, which are of particular interest to microbiologists.
Rethinking Culture
We are left with two competing truths: culturing is essential to the complete characterization of microbial communities and culturing cannot be scaled to analyze microbial communities.
The natural conclusion is that we must rethink our approach to culturing. Since spreading samples on a single agar plate is not the answer, the solution must lie in developing new technology that is capable of massively parallel isolation and cultivation of microbes. The ideal approach would separate microbes so that fast-growing members cannot overwhelm slower-growing strains. It would also be automated, with minimal human intervention required. Microarrays, with their thousands of wells, offer an idea of how an improved cultivation system might look. My company, for example, is developing a system that uses a high-density microfabricated array format to sort individual microbial strains into separate nanoscale growth chambers for cultivation.
The microbiome and metagenome results generated so far have been tantalizing, and suggest that there is much more to be gained if we can build on today’s genome-scale tools with a high-throughput system for isolating and cultivating the many members of a complex microbial community. Microbiologists around the world need innovative new tools to help them make sense of microbial populations in health, food, environmental, and other applications.
References
-
- Pérez-Jaramillo, J.E., Carrión, V.J., de Hollander, M. et al. The wild side of plant microbiomes. Microbiome 6, 143 (2018) doi:10.1186/s40168-018-0519-z microbiomejournal.biomedcentral.com/articles/10.1186/s40168-018-0519-z
- Berger, J.J. “Can Soil Microbes Slow Climate Change?” Scientific American. March 26, 2019. scientificamerican.com/article/can-soil-microbes-slow-climate-change/
- Fessler, J., Matson, V. & Gajewski, T.F. Exploring the emerging role of the microbiome in cancer immunotherapy. j. immunotherapy cancer 7, 108 (2019) doi: jitc.biomedcentral.com/articles/10.1186/s40425-019-0574-4
- Vivarelli, S., Salemi, R., Candido, S., Falzone, L., et al. (2019). Gut Microbiota and Cancer: From Pathogenesis to Therapy. Cancers, 11(1), 38. doi:10.3390/cancers11010038
Peter Christey, PhD, is the co-founder and CEO of General Automation Lab Technologies Inc.





Tell Us What You Think!