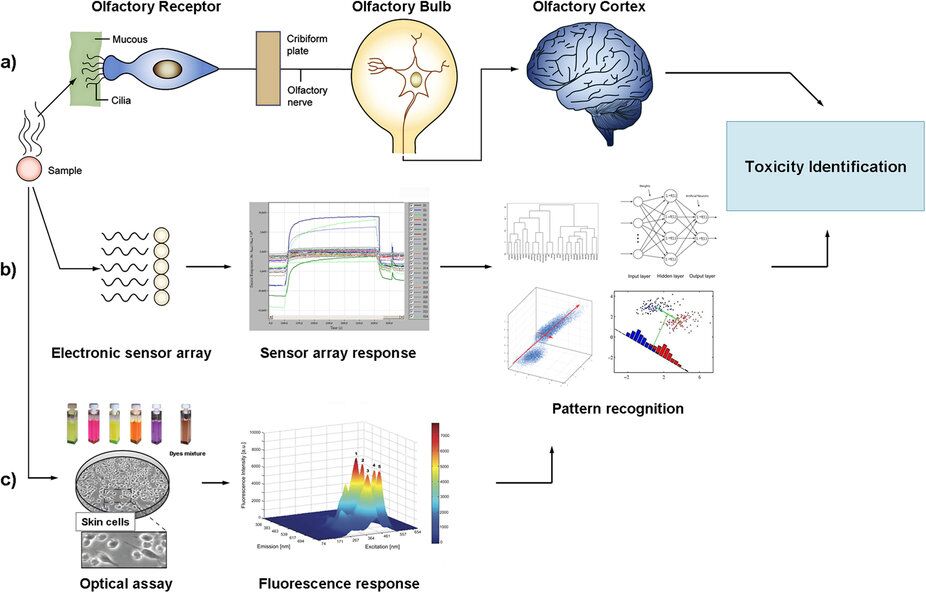
Schematic representation of sensing systems: (a) human olfactory system, (b) electronic analogue and (c) our optical analogue. (Credit: adapted with permission from E. Moczko, I. V. Meglinski, C. Bessant and S. A. Piletsky.Anal.Chem)
University of Leicester researchers have developed a new approach for analysing toxic chemicals in complex samples that mimics the way mammals smell and taste.
The technique could reduce the need for laboratory animals in biomedical research and other areas of chemical testing.
In the study a fluorescent assay combines a mixture of environmental-sensitive fluorescent dyes and human skin cells that generate fluorescence spectra patterns distinctive for particular physiological conditions.
Using multivariate data analysis, the optical signal is further processed, providing qualitative information and fast diagnostics.
The study was originally inspired by the operating principle of the electronic noses and tongues systems which mimic mammalian smell and taste recognition, combining semi-specific sensors and chemometric techniques for monitoring biochemical processes.
The Leicester Biotechnology Group at the University of Leicester has used similar principles, replacing electronic sensors with dyes array and applied them with human cells.
This combination has allowed transforming complex fluorescent spectra into a simple answer – whether the chemical compound is toxic or not.
While a digitized fluorescence image is a very high-dimensional vector – more than 250,000 numbers each – the number of tested chemicals is much less.
The researchers suggest the dimensionality reduction is the first task to overcome, with the challenge being how to transform the high dimension signal to a much lower dimension while keeping all important information safe.
Alexander Gorban, Professor of Applied Mathematics in the Department of Mathematics at the University of Leicester, said: “Firstly, we represented each signal by its projections on other signals. Secondly, we applied the classical and very popular model reduction method, Principal Component Analysis, and found five main components of the signals.
“Then we used dozens of various linear and nonlinear data analysis methods for the five-dimensional signals and validate the classifiers on the previously unseen data. Our approach can be considered as ‘explicit deep learning’, an explicit version of widely popularized (implicit) deep learning algorithms.”
The results had high accuracy, with both specificity and sensitivity above 90 per cent.
Sergey Piletsky, Professor of Bioanalytical Chemistry in the Department of Chemistry at the University of Leicester, commented: “The latest finding is a big step not only in toxicology, providing a modern, inexpensive and more efficient in vitro method but also in development of sensor assays for rapid quantification of a wide range of analytes which has always been a great challenge faced by analytical scientists.
“It can also reduce the need for laboratory animals in biomedical research, pharmaceutical industry, other areas of chemical testing and health diagnostics.”



