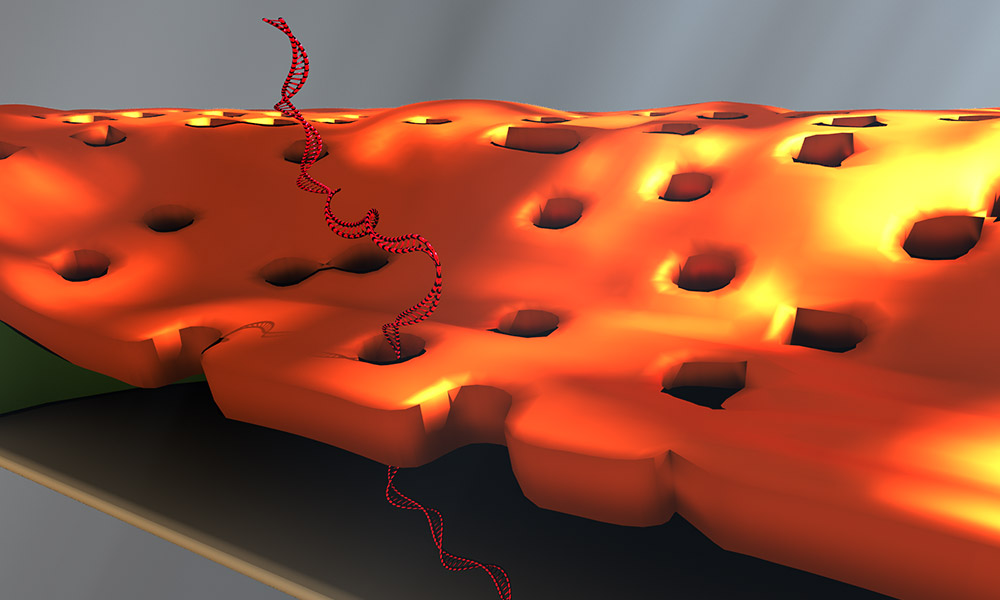
Doctoral student Greg Madejski’s illustration of the layers comprising his new DNA detection device. Image: University of Rochester illustration/Greg Madejski
Greg Madejski held his breath as he looked into the microscope, trying to weld two fingernail-sized chips together: a tiny chip containing a nanofilter on top of another chip with a DNA sensor.
It was frustrating work. The chips weren’t making good contact with each other. Madejski gently poked at the chips, then peered over the top of the microscope.
And exhaled.
The sudden waft of warm air swept over the nanofilter, transferring it to the sensor — right on target. The “accident” led Madejski to an important insight: the water vapor in his breath had condensed on the device, causing the nanofilter to adhere ever so neatly to the sensor.
“It was like a really high-tech temporary tattoo that I created by accident; lick and stick!” says the PhD student in the lab of James McGrath, a professor of biomedical engineering at the University of Rochester.
And that’s how water vapor became integral to the development and design of a novel device for detecting DNA biomarkers affiliated with disease. Created by McGrath’s lab in collaboration with Professor Vincent Tabard-Cossa and graduate student Kyle Briggs at the University of Ottawa, the device is described in an article published online at Nano Letters. The article, and an image from Madejski’s homemade animation of the device in operation, will be highlighted on the cover of the February 2018 print issue.
The device is comprised of three ultrathin layers:
- a nanoporous silicon nitride membrane which serves as a prefilter.
- a biosensor membrane with a single nanopore.
- a spacer layer that separates these by only 200 nm.
The arrangement creates a nanocavity filled with less than a femtoliter of fluid — or about a million times smaller than the smallest raindrops.
During operation, the device uses an electric field to lure a strand of DNA to enter one of the pores of the prefilter and then pass through the nanocavity to reach the pore of the underlying sensor membrane. This triggers changes in the device’s electrical current that can be detected and analyzed. The fact that DNA must elongate itself in a consistent way to pass through the two-membrane combination improves the precision and reproducibility of detection.
“This is a remarkable structure,” says McGrath. “We’ve built an integrated system with a highly porous filter within molecular reach of a sensor. I think there are many sensors, particularly those that hunt for biomarkers in raw biological fluids, that would benefit from filtering away unwanted molecules immediately upstream of the detector.”
The method of fabrication instantly wets the nanocavity, which is often difficult at the nanoscale. The device contains dozens of these nanocavities, which may eventually increase the amount of material that can be screened by enabling parallelized biomarker detection.
Tabard-Cossa’s lab uses solid-state nanopore devices to find new ways to manipulate and characterize single molecules. His lab was interested in finding new materials that could be used for biomarker detection. The prefilter in the new device addresses a problem with other silicon nanopore detectors: They are more likely to clog than alternative devices that use that biological pores for sensing. Biological membranes, on the other hand, are less stable than solid state nanopores, McGrath notes.
“We love to apply our membrane technologies to solve problems that others need solved. This is a very nice example,” McGrath says.
McGrath is co-founder of SiMPore, a University-based startup that develops highly portable, chip-based devices that incorporate silicon membranes for a variety of applications, from biological sensing to dialysis.
“I think we’re going to realize the practical advantages of this technology in the near term,” he says. A second generation of the new device, developed at SiMPore, incorporates the prefilter right on the chips during manufacturing at the wafer scale, “so there’s nobody breathing on it anymore,” he notes. “It’s actually all built as one unit and should make future studies very easy. That’s a credit to the ingenuity at SiMPore and quite a legacy for Greg.”
Source: University of Rochester




